Code
library(dplyr)
data(corrdat, package = "robumeta")
corrdat <-
corrdat %>%
distinct(studyid, esid, .keep_all = TRUE)James E. Pustejovsky
June 9, 2020
One common question about multivariate/multi-level meta-analysis is how such models assign weight to individual effect size estimates. When a version of the question came up recently on the R-sig-meta-analysis listserv, Dr. Wolfgang Viechtbauer offered a whole blog post in reply, demonstrating how weights work in simpler fixed effect and random effects meta-analysis and then how things get more complicated in multivariate models. I started thumb-typing my own reply as well, but then decided it would be better to write up a post so that I could use a bit of math notation (and to give my thumbs a break). So, in this post I’ll try to add some further intuition on how weights work in certain multivariate meta-analysis models. Most of the discussion will apply to models that include multiple level of random effects, but no predictors. I’ll also comment briefly on meta-regression models with only study-level predictor variables, and finally give some pointers to work on more complicated models.
It’s helpful to start by looking briefly at the basic fixed effect and random effects models, assuming that we’ve got a set of studies that each contribute a single effect size estimate so everything’s independent. Letting
In the basic random effects model, the weights for each study are proportional to
Now let’s consider the case where some or all studies in our synthesis contribute more than one effect size estimate. Say that we have effect sizes
There are many models that a meta-analyst might consider for this data structure. A fairly common one would be a model that includes random effects not only for between-study heterogeneity (as in the basic random effects model) but also random effects capturing within-study heterogeneity in true effect sizes. Let me write this model heirarchically, as
One thing to note about this model is that it treats all of the effect sizes as coming from a population with a common mean rma.mv() function from the metafor package to estimate it. I will acknowledge, though, that there will often be reason to use more complicated models, for example by replacing the overall average
To make some headway, it is helpful to first consider an even more specific model where, within a given study, all effect size estimates are equally precise and equally correlated. In particular, let’s assume that for each study
These assumptions might not be all that far-fetched. Within a given study, if the effect size estimates are for different measures of a common construct, it’s not unlikely that they would all be based on similar sample sizes (+/- a bit of item non-response). It might be a bit less likely if the effect size estimates are for treatment effects from different follow-up times (since drop-out/non-response tends to increase over time) or different treatment groups compared to a common control group—but still perhaps not entirely unreasonable. Further, it’s rather uncommon to have good information about the correlations between effect size estimates from a given study (because primary studies don’t often report all of the information needed to calculate these correlations). In practice, meta-analysts might need to simply make a rough guess about the correlations and then use robust variance estimation and/or sensitivity analysis to check themselves. And if we’re just ball-parking, then we’ll probably assume a single correlation for all of the studies.
The handy thing about this particular scenario is that, because all of the effect size estimates within a study are equally precise and equally correlated, the most efficient way to estimate an average effect for a given study is to just take the simple average (and, intuitively, this seems like the only sensible thing to do). To be precise, consider how we would estimate
Consider how precise each of the study-specific estimates are, relative to the true effects in their respective studies. Conditional on the true effect
There are several things worth noting about this expression for the weights. First, suppose that there is little between-study or within-study heterogeneity, so
Second, now suppose that there is no between-study heterogeneity (
Third and finally, between-study heterogeneity will tend to equalize the weights at the study level, so that the overall average is pulled closer to a simple average of the study-specific average effects. This works very much like in basic random effects meta-analysis, where increased heterogeneity will lead to weights that are closer to equal and an average effect size estimate that is closer to a simple average.
I think it’s useful to verify algebraic results like the ones I’ve given above by checking that you can reproduce them with real data. I’ll use the corrdat dataset from the robumeta package for illustration. The dataset has one duplicated row in it (I have no idea why!), which I’ll remove before analyzing further.
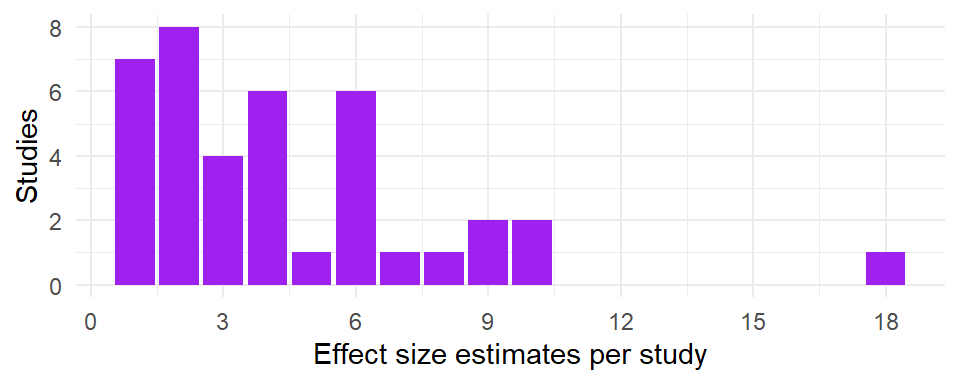
This dataset included a total of 171 effect size estimates from 39 unique studies. For each study, between 1 and 18 eligible effect size estimates were reported. Here is a histogram depicting the number of studies by the number of reported effect size estimates:
Here is the plot of the variances of each effect size versus the study IDs:
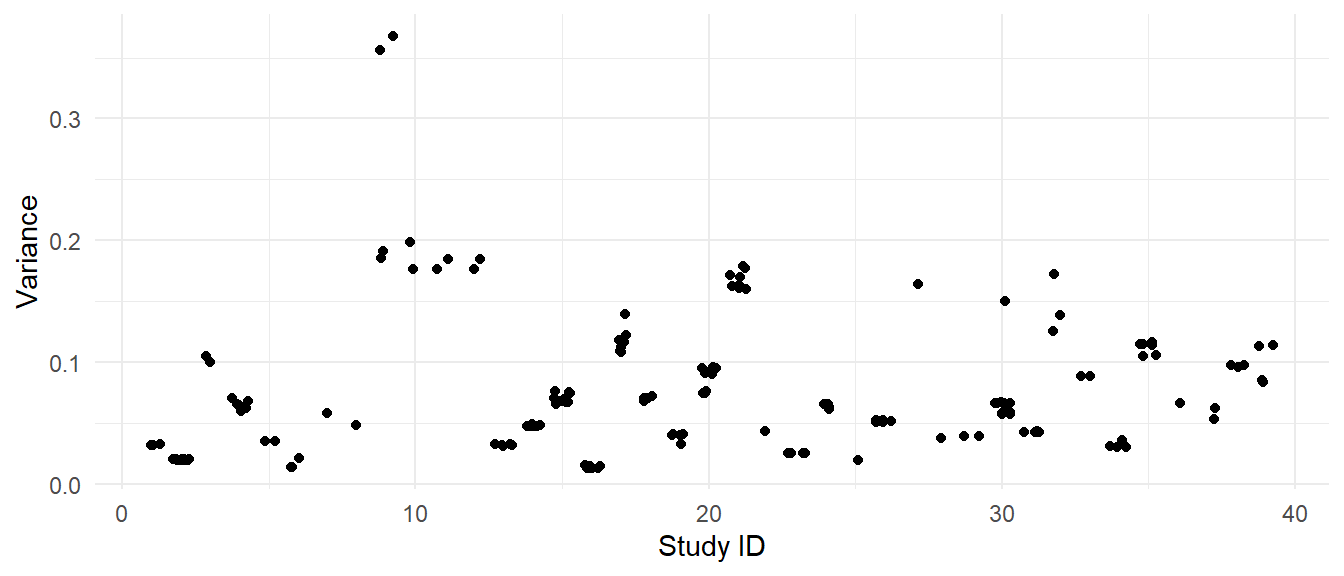
For most of the studies, the effect sizes have very similar sampling variances. One exception is study 9, where two of the effect sizes have variances of under 0.20 and the other two effect sizes have variances in excess of 0.35. Another exception is study 30, which has one effect size with much larger variance than the others.
Just for sake of illustration, I’m going to enforce my assumption that effect sizes have equal variances within each study by recomputing the sampling variances as the average sampling variance within each study. I will then impute a sampling variance-covariance matrix for the effect sizes, assuming a correlation of 0.7 for effects from the same study:
With this variance-covariance matrix, I can then estimate the multivariate meta-analysis model:
Multivariate Meta-Analysis Model (k = 171; method: REML)
logLik Deviance AIC BIC AICc
-94.7852 189.5703 195.5703 204.9777 195.7149
Variance Components:
estim sqrt nlvls fixed factor
sigma^2.1 0.0466 0.2159 39 no studyid
sigma^2.2 0.1098 0.3314 171 no studyid/esid
Test for Heterogeneity:
Q(df = 170) = 1141.4235, p-val < .0001
Model Results:
estimate se zval pval ci.lb ci.ub
0.2263 0.0589 3.8413 0.0001 0.1108 0.3417 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Based on this model, between-study heterogeneity is estimated as
I’ll first get the weights used in rma.mv to compute the overall average. The weights are represented as an
To verify that the formulas above are correct, I’ll use them to directly compute weights:
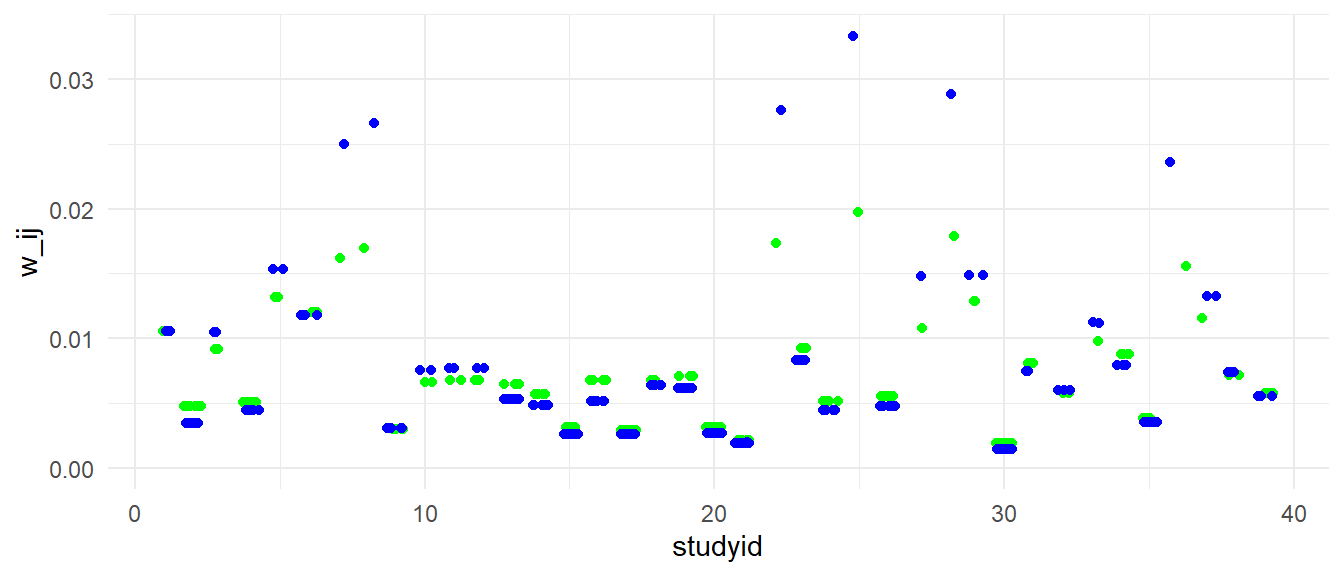
The weights I computed are perfectly correlated with the weights used rma.mv, as can be seen in the plot below.
If we remove the within-study random effect term from the model, the weights will be equivalent to setting
Multivariate Meta-Analysis Model (k = 171; method: REML)
Variance Components:
estim sqrt nlvls fixed factor
sigma^2 0.0951 0.3084 39 no studyid
Test for Heterogeneity:
Q(df = 170) = 1141.4235, p-val < .0001
Model Results:
estimate se zval pval ci.lb ci.ub
0.2235 0.0619 3.6122 0.0003 0.1022 0.3448 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Re-fitting the model with rma.mv() gives an between-study heterogeneity estimate of
[1] 0.2235231This matches the output of rma.mv().
Here is a plot showing the weights of individual effect sizes for each study. In blue are the weights under the assumption that
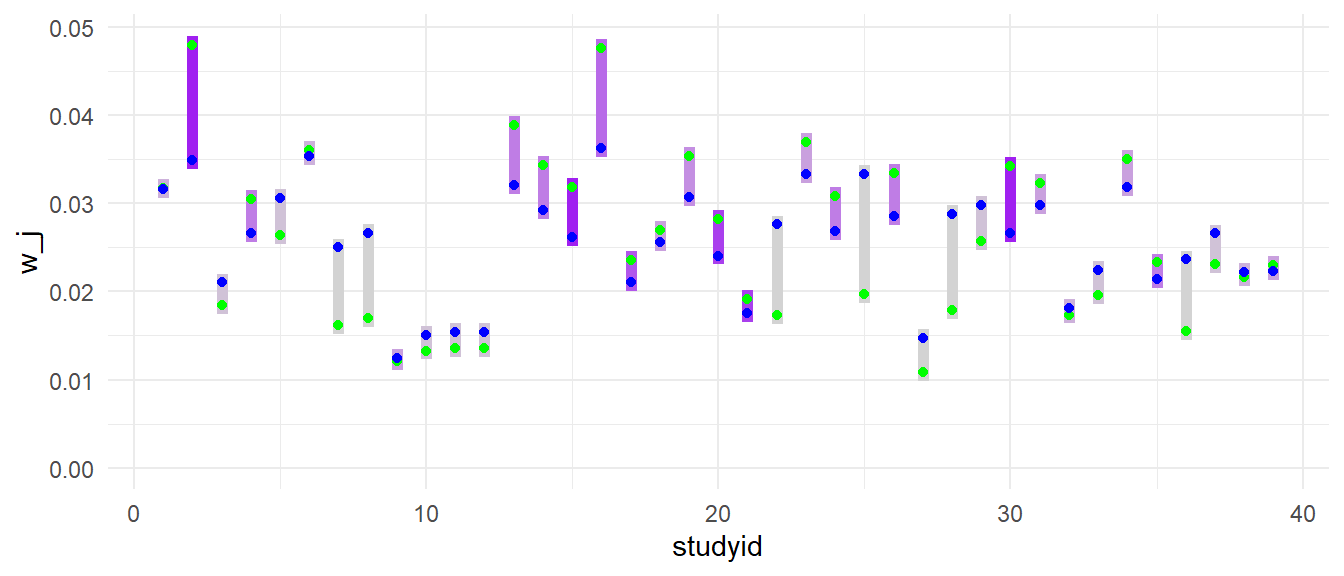
Below is a plot illustrating the changes in study-level weights (i.e., aggregating the weight assigned to each study). The bar color corresponds to the number of effect size estimates in each study; light grey studies have just one effect size, while studies with more effect sizes are more intensly purple. The notable drops in weight for studies with a single effect size estimate (light grey) are visible here too. Studies with more effect sizes (e.g., studies 2, 15, 30, with dark purple bars) gain weight when we allow
If we remove the restrictions that effect sizes from the same study have the same sampling variance and are equi-correlated, then the weights get a little bit more complicated. However, the general intuitions carry through. Let’s now consider the model with arbitrary sampling variance
One wrinkle with the more general form of the weights is that the effect-size level weights can sometimes be negative (i.e., negative
Some of the foregoing analysis also applies to models that include predictors. In particular, the formulas I’ve given for the weights will still hold for meta-regression models that include only study-level predictors. In other words, they work for models of the following form:
Here is an illustration with the corrdat meta-analysis. In these data, the variable college indicates whether the effect size comes from a college-age sample; it varies only at the study level. The variable males, binge, and followup have some within-study variation, which I’ll by taking the average of each of these predictors at the study level:
Now let’s fit a meta-regression model using all of the study-level predictors:
Multivariate Meta-Analysis Model (k = 171; method: REML)
logLik Deviance AIC BIC AICc
-86.6244 173.2488 187.2488 209.0327 187.9577
Variance Components:
estim sqrt nlvls fixed factor
sigma^2.1 0.0297 0.1723 39 no studyid
sigma^2.2 0.1068 0.3268 171 no studyid/esid
Test for Residual Heterogeneity:
QE(df = 166) = 1083.6655, p-val < .0001
Test of Moderators (coefficients 2:5):
QM(df = 4) = 13.0787, p-val = 0.0109
Model Results:
estimate se zval pval ci.lb ci.ub
intrcpt -0.0361 0.3678 -0.0982 0.9218 -0.7571 0.6849
college 0.2660 0.1384 1.9215 0.0547 -0.0053 0.5373 .
males_M 0.0023 0.0048 0.4753 0.6346 -0.0072 0.0118
binge_M 0.3441 0.1570 2.1927 0.0283 0.0365 0.6518 *
followup_M -0.0023 0.0011 -2.0379 0.0416 -0.0044 -0.0001 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1As you might expect, between-study heterogeneity is reduced a bit by the inclusion of these predictors.4
We can check my claim of computational equivalence by fitting the meta-regression model at the study level. Here I’ll aggregate everything up to the study level and compute the study-level weights:
tau_sq_reg <- MVMR_fit$sigma2[1]
omega_sq_reg <- MVMR_fit$sigma2[2]
corrdat_studylevel <-
corrdat %>%
group_by(studyid) %>%
mutate(n_j = n()) %>%
summarize_at(vars(effectsize, n_j, V_bar, college, binge_M, followup_M, males_M), mean
) %>%
mutate(
V_cond = (omega_sq_reg + (n_j - 1) * r * V_bar + V_bar) / n_j,
w_j = 1 / (tau_sq_reg + V_cond)
)Now I can fit a study-level meta-regression model. I use the weights argument to ensure that the meta-regression is estimated using the
Mixed-Effects Model (k = 39; tau^2 estimator: REML)
logLik deviance AIC BIC AICc
-13.0651 26.1303 38.1303 47.2884 41.2414
tau^2 (estimated amount of residual heterogeneity): 0.0297 (SE = 0.0264)
tau (square root of estimated tau^2 value): 0.1723
I^2 (residual heterogeneity / unaccounted variability): 26.89%
H^2 (unaccounted variability / sampling variability): 1.37
R^2 (amount of heterogeneity accounted for): 37.90%
Test for Residual Heterogeneity:
QE(df = 34) = 46.5050, p-val = 0.0748
Test of Moderators (coefficients 2:5):
QM(df = 4) = 13.0787, p-val = 0.0109
Model Results:
estimate se zval pval ci.lb ci.ub
intrcpt -0.0361 0.3678 -0.0982 0.9218 -0.7571 0.6849
college 0.2660 0.1384 1.9215 0.0547 -0.0053 0.5373 .
males_M 0.0023 0.0048 0.4753 0.6346 -0.0072 0.0118
binge_M 0.3441 0.1570 2.1927 0.0283 0.0365 0.6518 *
followup_M -0.0023 0.0011 -2.0379 0.0416 -0.0044 -0.0001 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The meta-regression coefficient estimates are essentially identical to those from the multi-variate meta-regression, although the between-study heterogeneity estimate differs slightly because it is based on maximizing the single-level model, conditional on an estimate of
In true multi-variate models, the meta-regression specification would typically include indicators for each dimension of the model. More generally, we might have a model that includes predictors varying within study, encoding characteristics of the outcome measures, sub-groups, or treatment conditions corresponding to each effect size estimate. The weights in these model get substantially more complicated, not in the least because the weights are specific to the predictors. For instance, in a model with four within-study predictors, a different set of weights is used in estimating the coefficients corresponding to each predictor. As Dr. Richard Riley noted on Twitter, relevant work on more complicated models includes this great paper by Dan Jackson and colleagues and this paper by Riley and colleagues. The latter paper demonstrates how multivariate models entail partial “borrowing of strength” across dimensions of the effect sizes, which is very helpful for building intuition about how these models work. I would encourage you to check out both papers if you are grappling with understanding how weights work in complex meta-regression models.
Note that this model also encompasses the multi-level meta-analysis described by Konstantopoulos (2011) and Van den Noortgate, et al. (2013) as a special case, with
Perhaps that makes sense, if you’ve carefully selected the set of effect sizes for inclusion in your meta-analysis. However, it seems to me that it could sometimes lead to perverse results. Say that all studies but one include just a single effect size estimate, each using the absolute gold standard approach to assessing the outcome, but that one study took a “kitchen sink” approach and assessed the outcome a bunch of different ways, including the gold standard plus a bunch of junky scales. Inclusion of the junky scales will lead to within-study heterogeneity, which in turn will pull the overall average effect size towards this study—the one with all the junk! That seems less than ideal, and the sort of situation where it would be better to select from the study with multiple outcomes the single effect size estimate based on the outcome assessment that most closely aligns with the other studies.↩︎
Things get even simpler if the model does not include within-study random effects, as I discussed in a previous post.↩︎
However, this need not be the case—it’s possible that introducing between-study predictors could increase the estimate of between-study heterogeneity. Yes, that’s totally counter-intuitive. Multi-level models can be weird.↩︎